Quality assessment description
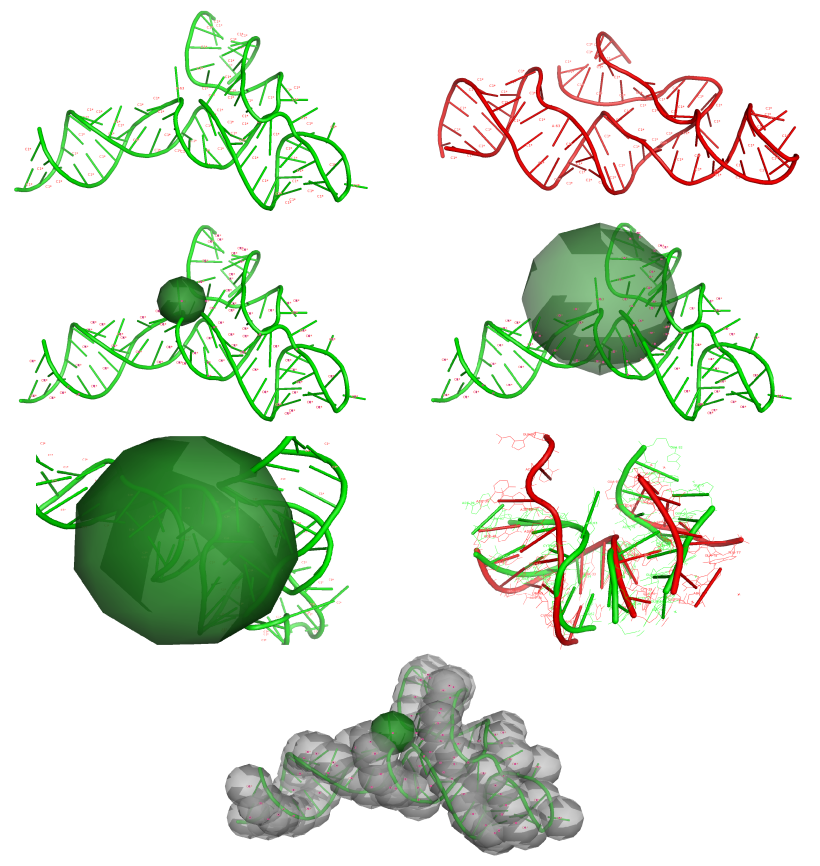
Figure 1. a) Input data defined by the user: the reference structure (in green), the model (in red), the center atom of the sphere, vector of radii, level of precision – positions of atoms of one type or all types will be analyzed; b) the sphere is built based on a selected atom from nucleotide for reference structure, the radius can be set by the user (e.g. 6 Å - left, 18 Å – right picture), c) atoms from the selected sphere are recognized, corresponding atoms from the model are identified, both sets are superimposed, and RMSD is calculated; d) spheres are built based on a selected type of atom for every nucleotide in the reference structure and the process from c) is repeated (reference structure: RNA Puzzles Problem 3, model: Dokholyan_model_2 (23)).
What you can do with the RNAlyzer tool?
RNAlyzer allows the user to evaluate the agreement between a predicted model of an RNA molecule and its reference structure by comparison of sets of atoms located inside a sphere. The sphere with a strictly defined radius is built around an atom selected by the user (e.g. C1*), which represents the center of the sphere of every nucleotide that exists in the reference structure. As a result of the sphere building process, a set of atoms that are included in a particular sphere is obtained. In the next stage of our method , for every sphere built on a reference structure, a corresponding set of atoms in the analyzed model is identified. Finally, the structures inside corresponding spheres are superimposed, and after that the RMSD value between corresponding sets is calculated. The ability to select different radii allows us to compare models at different levels of precision. If the value of a radius is small, then the model is analyzed from a local point of view (higher precision). Otherwise, the analysis is conducted from a global point of view (lower precision). If the sphere radius is equal to or greater than the radius of the whole molecule, the RMSD value is equal to the RMSD calculated for whole structure. Our software also allows the user to define a vector of sphere radii that can be visualized simultaneously in one plot.
System requirements
- » Java Platform, Standard Edition 6.0 http://java.com
- » Java 3D 1.5.2 https://java3d.java.net/binary-builds.html
- » OpenGL http://www.opengl.org/wiki/Getting_Started
How you can cite RNAlyzer?
Piotr Lukasiak, Maciej Antczak, Tomasz Ratajczak, Janusz M. Bujnicki, Marta Szachniuk, Mariusz Popenda, Ryszard W. Adamiak, Jacek Blazewicz, "RNAlyzer - novel approach for quality analysis of RNA structural models", Nucleic Acids Research, 2013, 41, pp. 5978-5990, (doi: 10.1093/nar/gkt318)